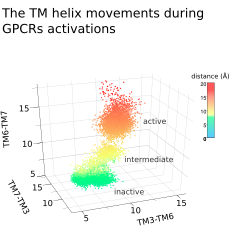
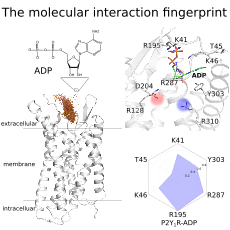
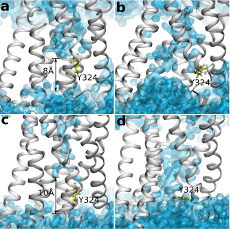
学术著作
* 通讯作者 #共同一作
2025
Conflitti, P.; Lyman, E.; Sansom, M. et al., Functional dynamics of G protein-coupled receptors reveal new routes for drug discovery. Nature Review Drug Discovery 2025
Dong J.; Wu C.; et al., Yuan, S.*, Enhancing Multifunctional Drug Screening via Artificial Intelligence. Digital Discovery 2025
Wu, C.; Li, Y.*; Vogel, H.*; Yuan, S.*, Rational Design Biased Compounds against μ-Opioid Receptor. ACS Pharmacology & Translational Science 2025
Xu, M.; Wu, C.; wang, S.; Zhan, W; Guo, L.; Li, Y; Vogel H; Yuan, S.*, Identifying Potent Compounds using Pairwise Consensus Methods. Journal of Chemical Information and Modeling 2025
2024
Introini, B.; Cui, W.; Chu, X.; Zhang, Y.; et al. Vogel, H.*; Yuan, S.*; Kudryashev, M.* Structure of Tetrameric Forms of the Serotonin-gated 5-HT3A Receptor Ion Channel. The EMBO Journal 2024.
Li, H.; Sun, X.; Cui, W.; Xu, M.; Dong J.; Ekundayo, BE; Ni, D.; Rao, Z.; Guo, L.; Stahlberg,H.*; Yuan, S.*; Vogel, H.*, Computational drug development for membrane protein targets. Nature Biotechnology 2024.
Wu, C.; Xu. M.; Dong, J.; Cui, W.; Yuan, S.*, The structure and function of olfactory receptors. Trends in Pharm Sci 2024.
Dong, J.; Wang, S.; Cui, W.; Sun, X.; Guo, H.; Yan H.; Vogel H.; Wang Z.*; Yuan, S.*, Machine learning deciphered molecular mechanistics with accurate kinetic and thermodynamic prediction. Journal of Chemical Theory and Computation 2024.
Zhang Q., Dong J., Zhou C., Zhang D., Yuan, S., Kramer D., Xue D., Peng C., Machine learning for data-driven design of high-safety lithium metal anode. STAR protocols 2024, 5 (1), 102834
Cui B., Pan Q., Cui W., Wang Y., Yuan, S., Loake G.*, Liu F.*, S-nitrosylation of a cytoplasmic receptor-like kinase regulates plant immunity. Science Advances 2024
Liu C.; Cui W.; et al. Inhibitor screening for volume-sensitive LRRC8A chloride channel. Journal of Biomolecular Structure and Dynamics 2024
2023
Cui, W.; Yuan, S.*, Will the hype of automated drug discovery finally be realized?. Expert Opinion on Drug Discovery 2023.
Yuan, Z.; Wang J.; Qu Q.; Zhu Z.; Xu M.; Zhao M.; Sun C.; Peng H.; Huang X.; Dong C.; Yuan, S.*; Li, Y.*, Celastrol Combats Methicillin-Resistant Staphylococcus aureus by Targeting Δ1-Pyrroline-5-Carboxylate Dehydrogenase. Advanced Science 2023.
Ma, T.; Wang, L.; Chai, A.; Liu, C.; Cui, W.; Yuan, S.; Wing Ngor Au, S.; Sun, L.; Zhang, X.; Zhang, Z.; Lu, J.; Gao, Y.; Wang, P.; Li, Z.; Liang, Y.; Vogel, H.*; Wang, Y. T.*; Wang, D.*; Yan, K.*; Zhang, H.*, Cryo-EM structures of ClC-2 chloride channel reveal the blocking mechanism of its specific inhibitor AK-42. Nat Commun 2023, 14 (1), 3424.
Zhang, H.; Wang, S.; Zhang, Z.; Hou, M.; Du, C.; Zhao, Z.; Vogel, H.; Li, Z.; Yan, K.; Zhang, X.; Lu, J.; Liang, Y.; Yuan, S.*; Wang, D.*; Zhang, H.*, Cryo-EM structure of human heptameric pannexin 2 channel. Nat Commun 2023, 14 (1), 1118.
Qu, Q.; Cui, W.; Huang, X.; Zhu, Z.; Dong, Y.; Yuan, Z.; Dong, C.; Zheng, Y.; Chen, X.; Yuan, S.*; Li, Y.*, Gallic Acid Restores the Sulfonamide Sensitivity of Multidrug-Resistant Streptococcus suis via Polypharmaceology Mechanism. J Agric Food Chem 2023, 71 (18), 6894-6907.
Cui, W.; Dong, J.; Wang, S.; Vogel, H.; Zou, R.; Yuan, S.*, Molecular basis of ligand selectivity for melatonin receptors. RSC Adv 2023, 13 (7), 4422-4430.
2022
Xu, M.; li, Y.; Dong, J.; Yuan, S.*, GPCR-IFP: A Web Server for Analyzing G-Protein Coupled ReceptorLigand Fingerprint Interaction from Experimental Structures. bioxrix 2022.
Xing, C.; Cui, W. Q.; Zhang, Y.; Zou, X. S.; Hao, J. Y.; Zheng, S. D.; Wang, T. T.; Wang, X. Z.; Wu, T.; Liu, Y. Y.; Chen, X. Y.; Yuan, S.* G.; Zhang, Z. Y.*; Li, Y. H.*, Ultrasound-assisted deep eutectic solvents extraction of glabridin and isoliquiritigenin from Glycyrrhiza glabra: Optimization, extraction mechanism and in vitro bioactivities. Ultrason Sonochem 2022, 83, 105946.
Wang, X.; Yuan, S.*; Chan, H. C. S.*, Translocation Mechanism of Allosteric Sodium Ions in beta(2)-Adrenoceptor. J Chem Inf Model 2022, 62 (12), 3090-3095.
Wang, S.; Sun, X.; Cui, W.; Yuan, S.*, MM/PB(GB)SA benchmarks on soluble proteins and membrane proteins. Front Pharmacol 2022, 13, 1018351.
Wang, H.; Wang, S.; Wang, J.; Shen, X.; Feng, X.; Yuan, S.; Sun, X.; Yuan, Q., Engineering a Prokaryotic Non-P450 Hydroxylase for 3'-Hydroxylation of Flavonoids. ACS Synth Biol 2022, 11 (11), 3865-3873.
Lan, D.; Li, S.; Tang, W.; Zhao, Z.; Luo, M.; Yuan, S.*; Xu, J.; Wang*, Y., Glycerol is Released from a New Path in MGL Lipase Catalytic Process. J Chem Inf Model 2022, 62 (9), 2248-2256.
Hou, Y.; Wang, S.; Bai, B.; Chan, H. C. S.*; Yuan, S.*, Accurate Physical Property Predictions via Deep Learning. Molecules 2022, 27 (5).
Chen, M.; Li, Y.; Li, S.; Cui, W.; Zhou, Y.; Qu, Q.; Che, R.; Li, L.; Yuan, S.*; Liu, X.*, Molecular Mechanism of Staphylococcus xylosus Resistance Against Tylosin and Florfenicol. Infect Drug Resist 2022, 15, 6165-6176.
2021
Zou, R.; Wang, X.; Li, S.; Chan, H. C. S.; Vogel, H.; Yuan, S.*, The role of metal ions in G protein‐coupled receptor signalling and drug discovery. WIREs Computational Molecular Science 2021, 12 (2).
Yao, D.; Wang, Y.; Zou, R.; Bian, K.; Yuan, S.*; Zhang, B.*; Wang, D.*, Wavelength-adjustable butterfly molecules in dynamic nanoassemblies for Extradomain-B fibronectin-modulating optical imaging and synchronous phototherapy of triple-negative breast cancer. Chemical Engineering Journal 2021, 420.
Xia, Y.; Zou, R.; Escouboue, M.; Zhong, L.; Zhu, C.; Pouzet, C.; Wu, X.; Wang, Y.; Lv, G.; Zhou, H.; Sun, P.; Ding, K.; Deslandes, L.; Yuan, S.*; Zhang, Z. M.*, Secondary-structure switch regulates the substrate binding of a YopJ family acetyltransferase. Nat Commun 2021, 12 (1), 5969.
Lan, D.; Zhao, G.; Holzmann, N.; Yuan, S.*; Wang, J.*; Wang, Y.*, Structure-Guided Rational Design of a Mono- and Diacylglycerol Lipase from Aspergillus oryzae: A Single Residue Mutant Increases the Hydrolysis Ability. J Agric Food Chem 2021, 69 (18), 5344-5352.
Cui, W.; Yu, F.; Zhang, Y.; Han, X.; Zou, R.; Tang, Y.; Wang, L.; Eliphaz, N.; Wang, J.; Yuan, S.*; Cai, X.; Li, Y., Discovery of traditional Chinese medicines against porcine reproductive and respiratory syndrome virus. Pharmacological Research - Modern Chinese Medicine 2021, 1.
Bai, B.; Zou, R.; Chan, H. C. S.; Li, H.*; Yuan, S.*, MolADI: A Web Server for Automatic Analysis of Protein-Small Molecule Dynamic Interactions. Molecules 2021, 26 (15).
2020
Liu, K.; Zou, R.; Cui, W.; Li, M.; Wang, X.; Dong, J.; Li, H.; Li, H.; Wang, P.; Shao, X.; Su, W.; Chan, H. C. S.; Li, H.; Yuan, S.*, Clinical HDAC Inhibitors Are Effective Drugs to Prevent the Entry of SARS-CoV2. ACS Pharmacol Transl Sci 2020, 3 (6), 1361-1370.
Liu, K.; Wu, L.; Yuan, S.; Wu, M.; Xu, Y.; Sun, Q.; Li, S.; Zhao, S.; Hua, T.; Liu, Z. J., Structural basis of CXC chemokine receptor 2 activation and signalling. Nature 2020, 585 (7823), 135-140.
Hua, T.; Li, X.; Wu, L.; Iliopoulos-Tsoutsouvas, C.; Wang, Y.; Wu, M.; Shen, L.; Brust, C. A.; Nikas, S. P.; Song, F.; Song, X.; Yuan, S.; Sun, Q.; Wu, Y.; Jiang, S.; Grim, T. W.; Benchama, O.; Stahl, E. L.; Zvonok, N.; Zhao, S.; Bohn, L. M.; Makriyannis, A.; Liu, Z.-J., Activation and Signaling Mechanism Revealed by Cannabinoid Receptor-Gi Complex Structures. Cell 2020, 180 (4), 655-665.e18.
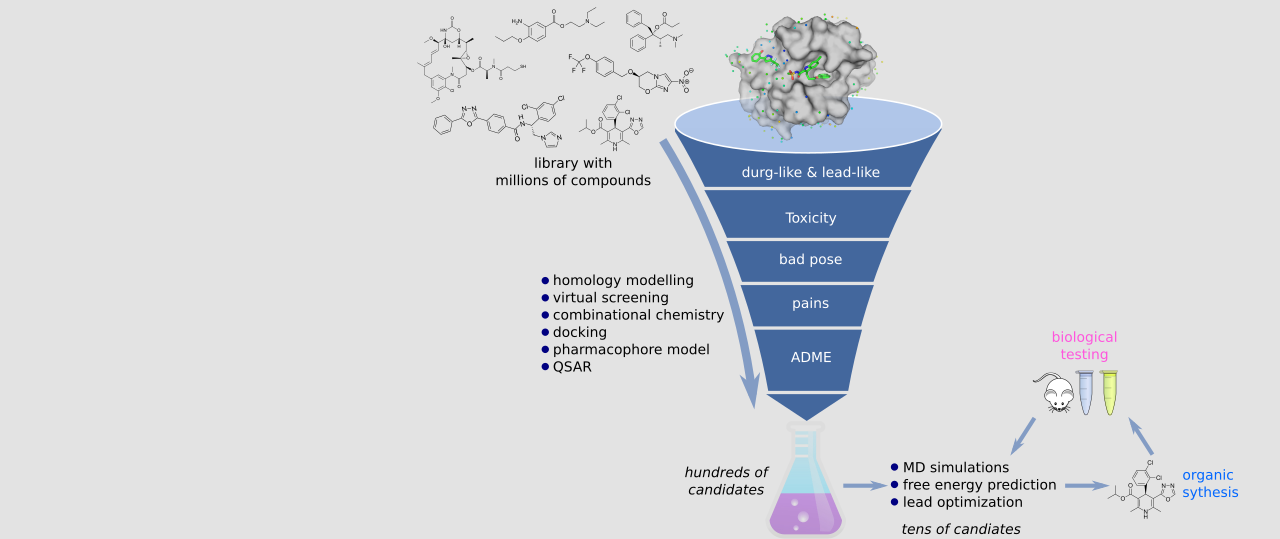
Cui, W.; Aouidate, A.; Wang, S.; Yu, Q.; Li, Y.; Yuan, S.*, Discovering Anti-Cancer Drugs via Computational Methods. Front Pharmacol 2020, 11, 733.
Chan, H. C. S.; Xu, Y.; Tan, L.; Vogel, H.; Cheng, J.*; Wu, D.*; Yuan, S.*, Enhancing the Signaling of GPCRs via Orthosteric Ions. ACS Cent Sci 2020, 6 (2), 274-282.
2019
Zheng, L.; Falschlunger, C.; Huang, K.; Mairhofer, E.; Yuan, S.; Wang, J.; Patel, D. J.; Micura, R.; Ren, A., Hatchet ribozyme structure and implications for cleavage mechanism. Proc Natl Acad Sci U S A 2019, 116 (22), 10783-10791.
Yuan, S.*; Dahoun, T.; Brugarolas, M.; Pick, H.; Filipek, S.; Vogel, H.*, Computational modeling of the olfactory receptor Olfr73 suggests a molecular basis for low potency of olfactory receptor-activating compounds. Commun Biol 2019, 2, 141.
Liu, L.; Chan, S.; Mo, T.; Ding, W.; Yu, S.; Zhang, Q.*; Yuan, S.*, Movements of the Substrate-Binding Clamp of Cypemycin Decarboxylase CypD. J Chem Inf Model 2019, 59 (6), 2924-2929.
Chen, X.; Xu, Y.; Qu, L.; Wu, L.; Han, G. W.; Guo, Y.; Wu, Y.; Zhou, Q.; Sun, Q.; Chu, C.; Yang, J.; Yang, L.; Wang, Q.; Yuan, S.; Wang, L.; Hu, T.; Tao, H.; Sun, Y.; Song, Y.; Hu, L.; Liu, Z. J.; Stevens, R. C.; Zhao, S.; Wu, D.; Zhong, G., Molecular Mechanism for Ligand Recognition and Subtype Selectivity of alpha(2C) Adrenergic Receptor. Cell Rep 2019, 29 (10), 2936-2943 e4.
Chan, H. C. S.; Shan, H.; Dahoun, T.; Vogel, H.; Yuan, S.*, Advancing Drug Discovery via Artificial Intelligence. Trends Pharmacol Sci 2019, 40 (8), 592-604.
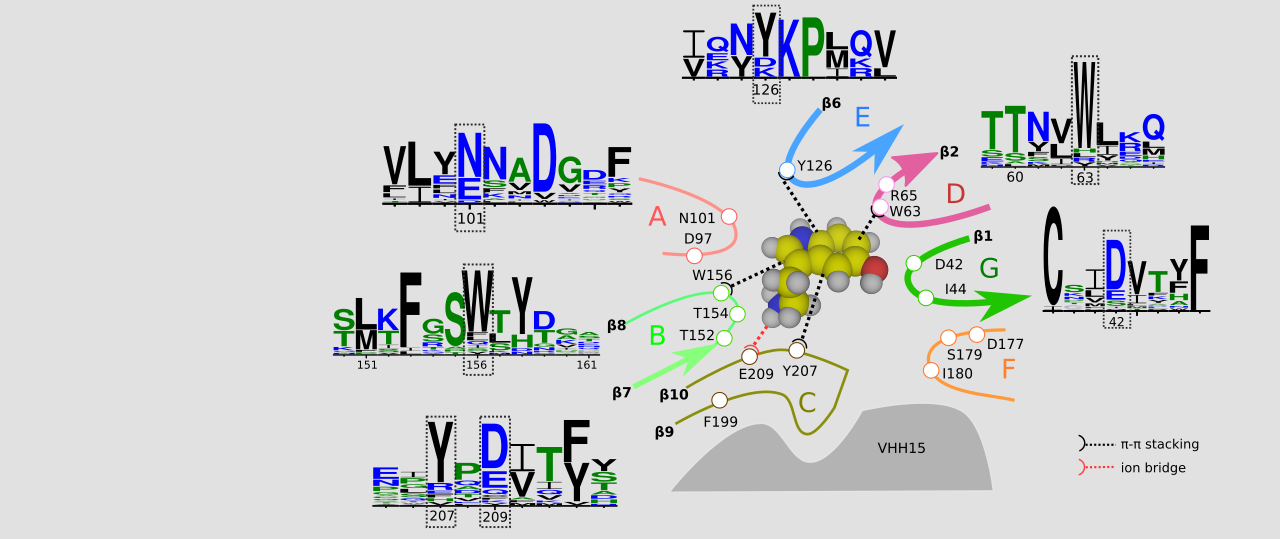
Chan, H. C. S.; Li, Y.; Dahoun, T.; Vogel, H.; Yuan, S.*, New Binding Sites, New Opportunities for GPCR Drug Discovery. Trends Biochem Sci 2019, 44 (4), 312-330.
2018
Zhou, Y.; Li, W.; You, W.; Di, Z.; Wang, M.; Zhou, H.; Yuan, S.; Wong, N. K.; Xiao, Y., Discovery of Arabidopsis UGT73C1 as a steviol-catalyzing UDP-glycosyltransferase with chemical probes. Chem Commun (Camb) 2018, 54 (52), 7179-7182.
Yan, Y.; Liu, Q.; Zang, X.; Yuan, S.; Bat-Erdene, U.; Nguyen, C.; Gan, J.; Zhou, J.; Jacobsen, S. E.; Tang, Y., Resistance-gene-directed discovery of a natural-product herbicide with a new mode of action. Nature 2018, 559 (7714), 415-418.
Sun, Z.; Wu, L.; Bocola, M.; Chan, H. C. S.; Lonsdale, R.; Kong, X. D.; Yuan, S.*; Zhou, J.*; Reetz, M. T.*, Structural and Computational Insight into the Catalytic Mechanism of Limonene Epoxide Hydrolase Mutants in Stereoselective Transformations. J Am Chem Soc 2018, 140 (1), 310-318.
Peng, Y.; McCorvy, J. D.; Harpsoe, K.; Lansu, K.; Yuan, S.; Popov, P.; Qu, L.; Pu, M.; Che, T.; Nikolajsen, L. F.; Huang, X. P.; Wu, Y.; Shen, L.; Bjorn-Yoshimoto, W. E.; Ding, K.; Wacker, D.; Han, G. W.; Cheng, J.; Katritch, V.; Jensen, A. A.; Hanson, M. A.; Zhao, S.; Gloriam, D. E.; Roth, B. L.; Stevens, R. C.; Liu, Z. J., 5-HT(2C) Receptor Structures Reveal the Structural Basis of GPCR Polypharmacology. Cell 2018, 172 (4), 719-730 e14.
Li, W.; Zhou, Y.; You, W.; Yang, M.; Ma, Y.; Wang, M.; Wang, Y.; Yuan, S.; Xiao, Y., Development of Photoaffinity Probe for the Discovery of Steviol Glycosides Biosynthesis Pathway in Stevia rebuadiana and Rapid Substrate Screening. ACS Chem Biol 2018, 13 (8), 1944-1949.
Ernst, I.; Haase, M.; Ernst, S.; Yuan, S.; Kuhn, A.; Leptihn, S., Large conformational changes of a highly dynamic pre-protein binding domain in SecA. Commun Biol 2018, 1, 130.
Chan, H. C. S.; Wang, J.; Palczewski, K.; Filipek, S.; Vogel, H.; Liu, Z. J.; Yuan, S.*, Exploring a new ligand binding site of G protein-coupled receptors. Chem Sci 2018, 9 (31), 6480-6489.
Chan, H. C. S.; Pan, L.; Li, Y.; Yuan, S.*, Rationalization of stereoselectivity in enzyme reactions. WIREs Computational Molecular Science 2018, 9 (4).
2017
Yuan, S.*; Chan, H. C. S.; Hu, Z., Using PyMOL as a platform for computational drug design. WIREs Computational Molecular Science 2017, 7 (2), e1298.
Yuan, S.*; Chan, H. C. S.; Hu, Z., Implementing WebGL and HTML5 in Macromolecular Visualization and Modern Computer-Aided Drug Design. Trends Biotechnol 2017, 35 (6), 559-571.
Yuan, H.; Zhang, J.; Cai, Y.; Wu, S.; Yang, K.; Chan, H. C. S.; Huang, W.; Jin, W. B.; Li, Y.; Yin, Y.; Igarashi, Y.; Yuan, S.*; Zhou, J.*; Tang, G. L.*, GyrI-like proteins catalyze cyclopropanoid hydrolysis to confer cellular protection. Nat Commun 2017, 8 (1), 1485.
Wang, J. B.; Ilie, A.; Yuan, S.*; Reetz, M. T.*, Investigating Substrate Scope and Enantioselectivity of a Defluorinase by a Stereochemical Probe. J Am Chem Soc 2017, 139 (32), 11241-11247.
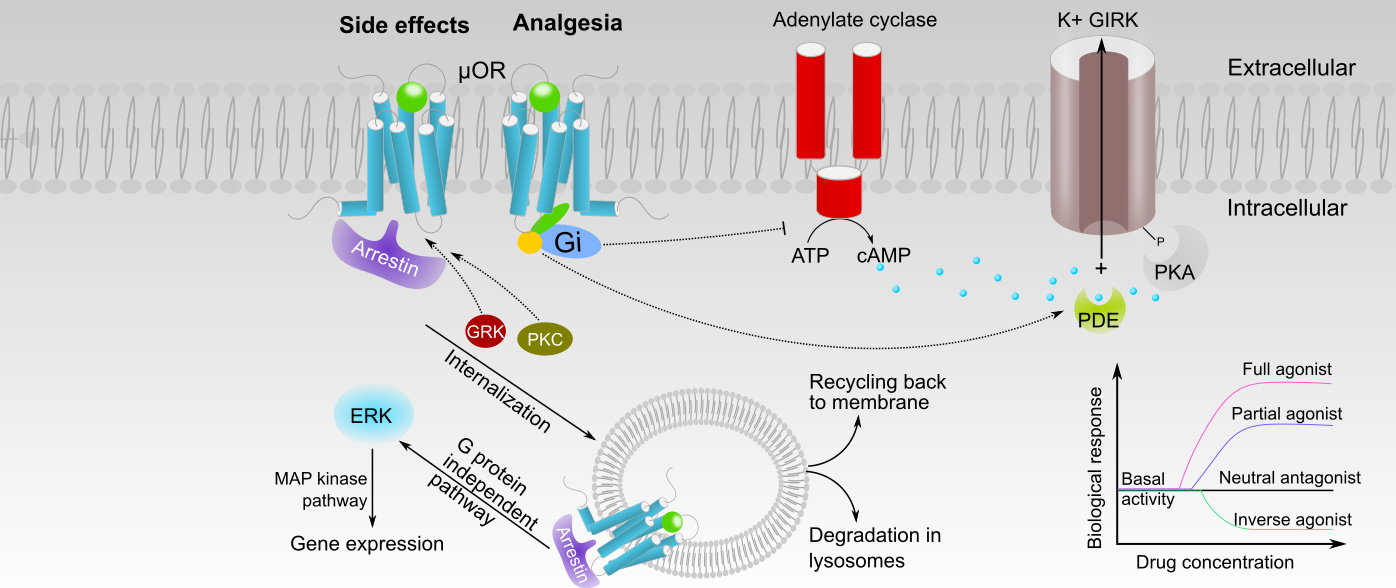
Chan, H. C. S.; McCarthy, D.; Li, J.; Palczewski, K.*; Yuan, S.*, Designing Safer Analgesics via mu-Opioid Receptor Pathways. Trends Pharmacol Sci 2017, 38 (11), 1016-1037.
2016
Zhang, Z. M.; Ma, K. W.; Yuan, S.; Luo, Y.; Jiang, S.; Hawara, E.; Pan, S.; Ma, W.; Song, J., Structure of a pathogen effector reveals the enzymatic mechanism of a novel acetyltransferase family. Nat Struct Mol Biol 2016, 23 (9), 847-52.
Yuan, S.*; Peng, Q.; Palczewski, K.; Vogel, H.; Filipek, S., Mechanistic Studies on the Stereoselectivity of the Serotonin 5-HT1A Receptor. Angew Chem Int Ed Engl 2016, 55 (30), 8661-5.
Yuan, S.*; Filipek, S.; Vogel, H., A Gating Mechanism of the Serotonin 5-HT3 Receptor. Structure 2016, 24 (5), 816-825.
Yuan, S.*; Chan, H. C. S.; Filipek, S.; Vogel, H., PyMOL and Inkscape Bridge the Data and the Data Visualization. Structure 2016, 24 (12), 2041-2042.
Yuan, S.*; Chan, H. C.; Vogel, H.; Filipek, S.; Stevens, R. C.; Palczewski, K., The Molecular Mechanism of P2Y1 Receptor Activation. Angew Chem Int Ed Engl 2016, 55 (35), 10331-5.
Ji, X.; Liu, W. Q.; Yuan, S.; Yin, Y.; Ding, W.; Zhang, Q., Mechanistic study of the radical SAM-dependent amine dehydrogenation reactions. Chem Commun (Camb) 2016, 52 (69), 10555-8.
Chan, H. C.; Filipek, S.; Yuan, S.*, The Principles of Ligand Specificity on beta-2-adrenergic receptor. Sci Rep 2016, 6, 34736.
2015
Yuan, S.*; Palczewski, K.; Peng, Q.; Kolinski, M.; Vogel, H.; Filipek, S., The mechanism of ligand-induced activation or inhibition of mu- and kappa-opioid receptors. Angew Chem Int Ed Engl 2015, 54 (26), 7560-3.
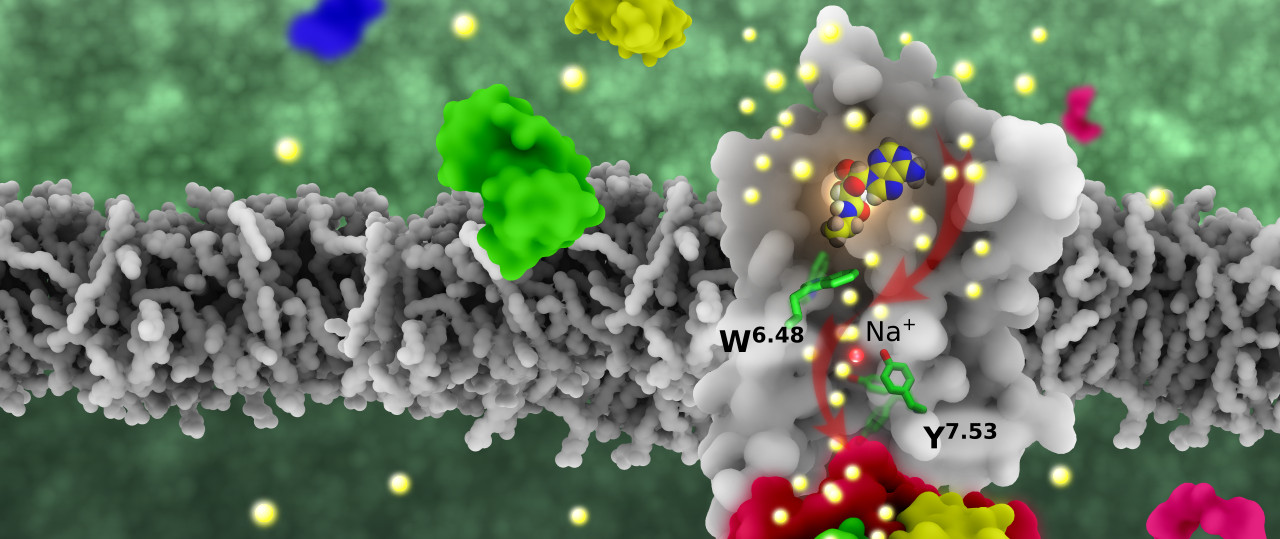
Yuan, S.*; Hu, Z.; Filipek, S.; Vogel, H., W246(6.48) opens a gate for a continuous intrinsic water pathway during activation of the adenosine A2A receptor. Angew Chem Int Ed Engl 2015, 54 (2), 556-9.
Baud, O.#; Yuan, S.#; Veya, L.; Filipek, S.; Vogel, H.; Pick, H.*, Exchanging ligand-binding specificity between a pair of mouse olfactory receptor paralogs reveals odorant recognition principles. Sci Rep 2015, 5, 14948.
2014
Yuan, S.*; Filipek, S.; Palczewski, K.; Vogel, H.*, Activation of G-protein-coupled receptors correlates with the formation of a continuous internal water pathway. Nat Commun 2014, 5, 4733.
Kufareva, I.; Katritch, V.; Participants of, G. D.; Stevens, R. C.; Abagyan, R., Advances in GPCR modeling evaluated by the GPCR Dock 2013 assessment: meeting new challenges. Structure 2014, 22 (8), 1120-1139.
Kong, X. D.; Yuan, S.; Li, L.; Chen, S.; Xu, J. H.; Zhou, J., Engineering of an epoxide hydrolase for efficient bioresolution of bulky pharmaco substrates. Proc Natl Acad Sci U S A 2014, 111 (44), 15717-22.
2013
Yuan, S.*; Vogel, H.; Filipek, S.*, The role of water and sodium ions in the activation of the mu-opioid receptor. Angew Chem Int Ed Engl 2013, 52 (38), 10112-5.
Yuan, S.*; Wu, R.; Latek, D.; Trzaskowski, B.; Filipek, S.*, Lipid receptor S1P(1) activation scheme concluded from microsecond all-atom molecular dynamics simulations. PLoS Comput Biol 2013, 9 (10), e1003261.
Wu, Y.; Yuan, S.; Chen, S.; Wu, D.; Chen, J.; Wu, J., Enhancing the production of galacto-oligosaccharides by mutagenesis of Sulfolobus solfataricus beta-galactosidase. Food Chem 2013, 138 (2-3), 1588-95.
before 2013
Le Roy, K.; Vergauwen, R.; Struyf, T.; Yuan, S.; Lammens, W.; Matrai, J.; De Maeyer, M.; Van den Ende, W., Understanding the role of defective invertases in plants: tobacco Nin88 fails to degrade sucrose. Plant Physiol 2013, 161 (4), 1670-81.
Yuan, S.; Le Roy, K.; Venken, T.; Lammens, W.; Van den Ende, W.; De Maeyer, M., pKa modulation of the acid/base catalyst within GH32 and GH68: a role in substrate/inhibitor specificity? PLoS One 2012, 7 (5), e37453.
Yuan, S.; Ghoshdastider, U.; Trzaskowski, B.; Latek, D.; Debinski, A.; Pulawski, W.; Wu, R.; Gerke, V.; Filipek, S., The role of water in activation mechanism of human N-formyl peptide receptor 1 (FPR1) based on molecular dynamics simulations. PLoS One 2012, 7 (11), e47114.
Trzaskowski, B.; Latek, D.; Yuan, S.; Ghoshdastider, U.; Debinski, A.; Filipek, S., Action of molecular switches in GPCRs--theoretical and experimental studies. Curr Med Chem 2012, 19 (8), 1090-109.
Lammens, W.; Le Roy, K.; Yuan, S.; Vergauwen, R.; Rabijns, A.; Van Laere, A.; Strelkov, S. V.; Van den Ende, W., Crystal structure of 6-SST/6-SFT from Pachysandra terminalis, a plant fructan biosynthesizing enzyme in complex with its acceptor substrate 6-kestose. Plant J 2012, 70 (2), 205-19.
Zheng, C.; Liu, H. H.; Yuan, S.; Zhou, J.; Zhang, B., Molecular basis of LMAN1 in coordinating LMAN1-MCFD2 cargo receptor formation and ER-to-Golgi transport of FV/FVIII. Blood 2010, 116 (25), 5698-706.


 袁曙光博士2009年于中科院上海有机化学研究所(SIOC)取得结构生物学方向硕士学位。之后,他获得欧盟玛丽居里全额奖学金资助,先后在比利时鲁汶大学(KULeuven),波兰科学院和洛桑瑞士联邦理工学院(EPFL)共同完成博士学习,主攻方向为计算生物学与计算机辅助药物设计。2013年,他获得最佳博士论文奖。毕业后,他荣获玛丽居里ETH博士后奖学金。2014年,袁博士创造性提出GPCRs的激活机制与连续水分子通路(Nature Communications,2014),并得到同行的广泛认可。此后袁博士在瑞士一家领头制药公司从事计算机辅助药物设计(CADD)工作5年时间。期间,袁博士推进两个GPCR药物分子已经进入临床测试阶段,并且获批国际专利。其中1个分子于2022年初被FDA所批准,另一个分子进入临床二期。2018年,袁博士被波兰科学院授予“荣誉教授”称号。2019年,袁博士创立阿尔法分子科技有限责任公司,专注GPCR新药研发。2022年,成功入选全球“top 2%”顶尖科学家榜单。
年被中国药科大学聘为产业教授。
袁曙光博士2009年于中科院上海有机化学研究所(SIOC)取得结构生物学方向硕士学位。之后,他获得欧盟玛丽居里全额奖学金资助,先后在比利时鲁汶大学(KULeuven),波兰科学院和洛桑瑞士联邦理工学院(EPFL)共同完成博士学习,主攻方向为计算生物学与计算机辅助药物设计。2013年,他获得最佳博士论文奖。毕业后,他荣获玛丽居里ETH博士后奖学金。2014年,袁博士创造性提出GPCRs的激活机制与连续水分子通路(Nature Communications,2014),并得到同行的广泛认可。此后袁博士在瑞士一家领头制药公司从事计算机辅助药物设计(CADD)工作5年时间。期间,袁博士推进两个GPCR药物分子已经进入临床测试阶段,并且获批国际专利。其中1个分子于2022年初被FDA所批准,另一个分子进入临床二期。2018年,袁博士被波兰科学院授予“荣誉教授”称号。2019年,袁博士创立阿尔法分子科技有限责任公司,专注GPCR新药研发。2022年,成功入选全球“top 2%”顶尖科学家榜单。
年被中国药科大学聘为产业教授。